HT-SAXS Instructions the basics
Introduction Registration Samples Spreadsheet Shipping Data Resources CitingIntroduction mail-in HT-SAXS at SIBYLS
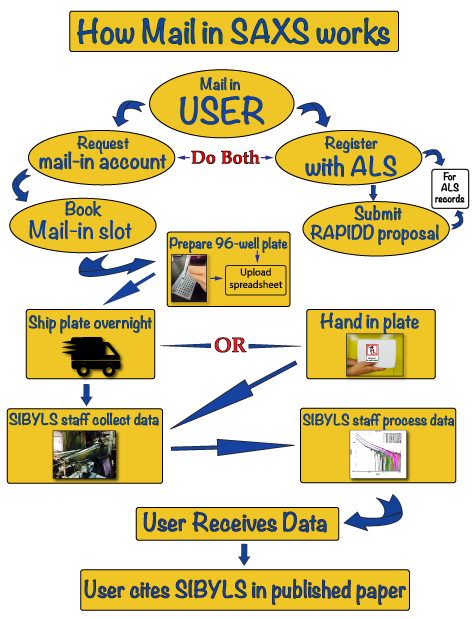
HT-SAXS workflow
Below is a schematic diagram of the High Throughput Small Angle X-ray Scattering (HT-SAXS) workflow that you may take advantage of at the SIBYLS beamline 12.3.1 at the Advanced Light Source in Berkeley:
Robotics and detector
To achieve sufficient X-ray flux for informative scattering with low protein concentration and small volumes, we designed the SIBYLS beamline at the Advanced Light Source. We employ a light path generated by a super-bend magnet to provide a 1012 photons/sec flux (1 Å wavelength). The tunable incident wavelength enables rapid adjustment of the q range appropriate for the experiment without changing the sample to detector configuration (q=4π sin(θ /2)/λ where θ is the scattering angle and λ is the wavelength).
To make SIBYLS truly high-throughput, we installed a new Pilatus3 2M pixel array detector from Dectris, which gives much faster readout times over our previous MAR165 area detector.

SIBYLS then installed a new Tecan Freedom Evo liquid handling robot that divides the time it takes to collect a full plate in half, from approximately 5 hours to 2.5 hours per 96-well plate (with the potential to collect a plate in 10 minutes over the coming year).
We believe this will be a game changer for many reasons. We will be able to accommodate more users per week, enable new screening methods, and have more time to offer SEC-coupled SAXS and time resolved SAXS.
In addition to the new robotics, we have improved instruments for higher data quality. Our new monitor of transmitted beam intensity should greatly reduce buffer subtraction errors. We have also replaced the windows for lower background. In completing these upgrades, the stage is now set for increased sample to detector distance providing access to lower q (an upgrade that will occur sometime during the summer).
Registration request beamtime using these 4 steps
1. Request mail-in account
All SIBYLS MailinSAXS users must first register for a MailinSAXS account
Request Mail-in Account2. Register with ALS & Submit a RAPIDD Proposal
All users must register with the ALS User Office, and submit one ALS RAPIDD proposal for every three MailinSAXS slots
3. Register at Simple Scattering
To expedite processing and sending your data, your data will be sent to Simple Scattering. Please register by clicking the button below
Simple Scattering4. Book mail-in slot
After completing the previous 3 steps, you may book a mail-in slot for beamtime
Book Mail-in SlotSamples information on plate and sample requirements
Prepare 96-well plate
Our sampling robot uses specific 96-well Full-skirt Clear (not opaque) PCR Microplates. You must use this particular plate for your samples to be collected. We recommend the AXYGEN brand from Corning. Here is a link to order plates from Corning: Order Plates
You will also need to use the AxyMat Sealing Mat for 96-well PCR Microplates to minimize evaporation. Here is a link to order mats: Order Mats
Sample requirements
Fill plate wells with a consistent volume of 30µL of sample and buffer. It is extremely important to keep the volume consistent throughout the plate.
Three sample concentrations between 1-10 mg/ml.
For redundancy, it is recommended to collect two (identical) buffers for each concentration series, one before and one after.
Do not use greater than 2% glycerol. It has been shown that 2% provides the maximum benefit. With glycerol, the viscosity of the solution increases and bubbles can become a problem. See our Methods paper for more information.
For more information, see SAXS Sample Prep
Also see the SIBYLS Mail-in HT-SAXS tutorial video on our WordPress site
Spreadsheet sample entry and template
Upload spreadsheet
Our robot uses a spreadsheet to determine how to run your samples. Please follow the next four steps to stage your samples for data collection:
1. Download the spreadsheet template by clicking the button below:
Download Template
2. Fill it out as follows:
-
Column A
ContainerID. "LabName". Write the name of your lab name here (no spaces).
-
Column B
ContainerType. "plate". Do not change this column.
-
Column C
Well. Indicates well location. Do not change any value in this column.
-
Column D
SampleID. Write the name for the data output file (Sample ID). Only letters, numbers and underscores are accepted (no spaces).
-
Column E
Collect. When checked ("x"), the sample will be collected (in order, starting from the top). If there is no sample or buffer in the well, leave it blank.
-
Column F
Directory. Each sample will have it's own directory named after the well number. For example "A1", "A2", etc. If there is buffer in the well, add a "b" after the well number. This is very important for accurate data processing.
-
Column G
WashAfter. Indicates a wash step. When checked, the sample cell will be thoroughly rinsed three times after that well is exposed. It is recommended to wash after a concentration series.
| A | B | C | D | E | F | G |
|---|---|---|---|---|---|---|
| ContainerID | ContainerType | Well | SampleID | Collect | Directory | WashAfter |
| LabName | plate | A1 | buffer1 | x | A1b | |
| LabName | plate | A2 | protein_1_low | x | A2 | |
| LabName | plate | A3 | protein_1_medium | x | A3 | |
| LabName | plate | A4 | protein_1_high | x | A4 | x |
| LabName | plate | A5 | buffer1 | x | A5b | |
| LabName | plate | A6 | buffer2 | x | A6b | |
| LabName | plate | A7 | protein_2_low | x | A7 | |
| LabName | plate | A8 | protein_2_medium | x | A8 | |
| LabName | plate | A9 | protein_2_high | x | A9 | x |
| LabName | plate | A10 | buffer2 | x | A10b | |
| LabName | plate | A11 | buffer3 | x | A11b | |
| LabName | plate | A12 | protein_3_low | x | A12 | |
| LabName | plate | B1 | protein_3_medium | x | B1 | |
| LabName | plate | B2 | protein_3_high | x | B2 | x |
| LabName | plate | B3 | buffer3 | x | B3b | |
| LabName | plate | B4 | B4 (no sample) |
3. Upload your updated spreadheet to the SIBYLS SAXS database
Upload Spreadsheet4. Once you have booked a mail-in slot, a unique barcode will be created for your plate. Print this label out and tape it to the side of your plate.
Example Label:
Shipping how to package and ship your plates
Ship plate overnight
-
How to Safely Pack Your Plate:
- Seal plate securely with recommended sealing mat. DO NOT use adhesive based sealing film. Be sure label is securely taped to side of plate.
- Place sealed plate in plastic bag or wrap with parafilm.
- Using two 4°C cold packs (NOT frozen), place one on top and one on bottom of plate.
- Rubberband the assembly together for placement within styrofoam shipping box.
- Tighty pack styrofoam box by surrounding assembly with more cold packs.
- Okay if these are frozen, but do not allow plate to touch any frozen packs. DO NOT ship with wet ice.
- Clearly label the outside of your shipping box with the temperature of your samples. This is extremely important if a shipping delay occurs.
- Provide us with a tracking number
Shipping address
Ship samples overnight to arrive at SIBYLS on the first day of your shift:
- Lawrence Berkeley Lab
- ATTN: Kathryn Burnett / SIBYLS 12.3.1
- 1 Cyclotron Road MS 6R2136
- Berkeley, CA 94720
- Phone: 510-685-5755
SHIPPING ADDRESS:
Data what happens when and after your plate arrives
SIBYLS staff collect data
-
The following outlines how your samples will be processed:
- You will be notified by email that your samples have arrived.
- The plate will be stored at the appropriate temperature: 4°C or -80°C.
- It will be lightly spun at 3700 rpm for 10 minutes at 4°C prior to data collection.
- It will be held at 10°C during data collection, which lasts approximately 2.5 hours for a full plate.
- You will be notified by email as soon as your data is available.
SIBYLS staff process data
You will get back integrated scattering profiles in ASCI format in a results directory for each sample. A series of exposures, in equal sub-second time slices, are taken of each well.
In addition, you will receive a report with an initial assessment of the sample from our beamline staff.
Accessing and transferring your data
You will receive an email once the data package is available. The results will be stored and accessible in your data directory i.e. /data/username/ on our kona server. Where username is your individual or lab's login name.
To access data in your remote SIBYLS directory open a Terminal window and log into our server kona
local>ssh username@kona.als.lbl.gov
Then once logged in change directories to your data directory and list the contents:
[username@kona ~]$ cd /data/username
[username@kona ~]$ ls
To transfer data to your local computer, open a Terminal window and make or navigate to target directory to deposit your results or data. Example:
local> mkdir saxs
local> cd saxs
Then transfer the files using either remote sync rsync or secure copy scp, for example:
local > rsync -avz username@kona.als.lbl.gov:/data/username/20180101_username_Results .
would copy over the entire 20180101_username_Results directory from our server to your local machine using rsync and the "kona" server.
An example of just copying over your .dat files using scp and the "kona" server would be:
local>scp -rp username@kona.als.lbl.gov:/data/username/20180101_username_Results/*.dat .
Type man rsync or man scp in your Terminal for more information.
You may also use sftp or other GUI-based tools to transfer data to your local computer.
This is helpful in particular if you want to analyze your data with tools such as SAXS Frameslice, SAXS Similarity, or our other software.
If you have any questions, please contact Kathryn.
Citing the key to keep SIBYLS running to serve you
Cite SIBYLS in published papers
Please cite all relevant SIBYLS beamline papers below in the 'References' section of your journal article or book chapter
- Rosenberg DJ, Hura GL, Hammel M (2022) "Size exclusion chromatography coupled small angle X-ray scattering with tandem multiangle light scattering at the SIBYLS beamline" Meth. enzymol. 677, 191-219. (SEC-SAXS)
- Classen S, et al. (2013) “Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source” J. Appl. Crystallogr. 46(Pt 1), 1-13. (SIBYLS Beamline hardware)
- Putnam CD, Hammel M, Hura GL, Tainer JA (2007) "Solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution” Q. Rev. Biophys. 40, 191-285. (General SAXS)
Acknowledge SIBYLS
Please also acknowledge SIBYLS when using data generated at beamline 12.3.1.
These citations and acknowledgments are absolutely essential in keeping the MailinSAXS program funded and the beamline continually repaired and upgraded, as well as to provide access to experienced staff and the development of new technologies, such as SEC-SAXS.
Resources where to get additional information
Feedback
We are continuing to improve our systems with the aim of making data of higher quality. Please provide us with constructive feedback. For additional information please feel free to contact:
- Kathryn Burnett
- Research Associate
- kburnett@lbl.gov
- Greg Hura, Ph.D
- SAXS Scientist
- glhura@lbl.gov
Additional details
Learn about common problems with SAXS samples:
SAXS StatisticsOnline Webinars
We regularly present online webinars to highlight new SAXS features, and how to address and solve specific problems that occur during SAXS data collection and processing:
Webinars